Introduction
Proteins are the centre of many studies and used in a huge variety of applications in both industry and academia, ranging from the fields of chemistry, biology to medicine and agriculture. Accordingly, there is great need for the rapid and efficient identification of protein variants with new and improved properties.1
Traditional workflows for the evaluation of novel protein variants comprise DNA assembly, its transformation into a heterologous host as well as the production, purification, and analysis of the resulting product.2 Here, most frequently site-saturation mutagenesis is employed, which uses degenerated primers in a PCR for the targeted introduction of mutations. Downsides such as the risk for unwanted expansion of gene sequences and the dependence on elaborate cell lysates or cellular protein synthesis and purification however can impair the functionality of libraries and make this method quite time-consuming and labour intensive.3
Expression systems consisting of rolling circle amplification (RCA) and protein synthesis in cell-free extracts can be used to circumvent these hurdles. However, their potential has not been fully explored yet due to the requirement of high total reaction volumes of 15 µL, long execution times of at least 12 h, a high number of workflow steps and changing of reaction vessels. This application note highlights HyperXpress, a novel approach which allows to perform RCA and protein synthesis in cell-free extracts in a faster, low-volume single vessel reaction. As a proof of principle this approach was then used to identify new-to nature sequence variants of GFP.4
Assay principle
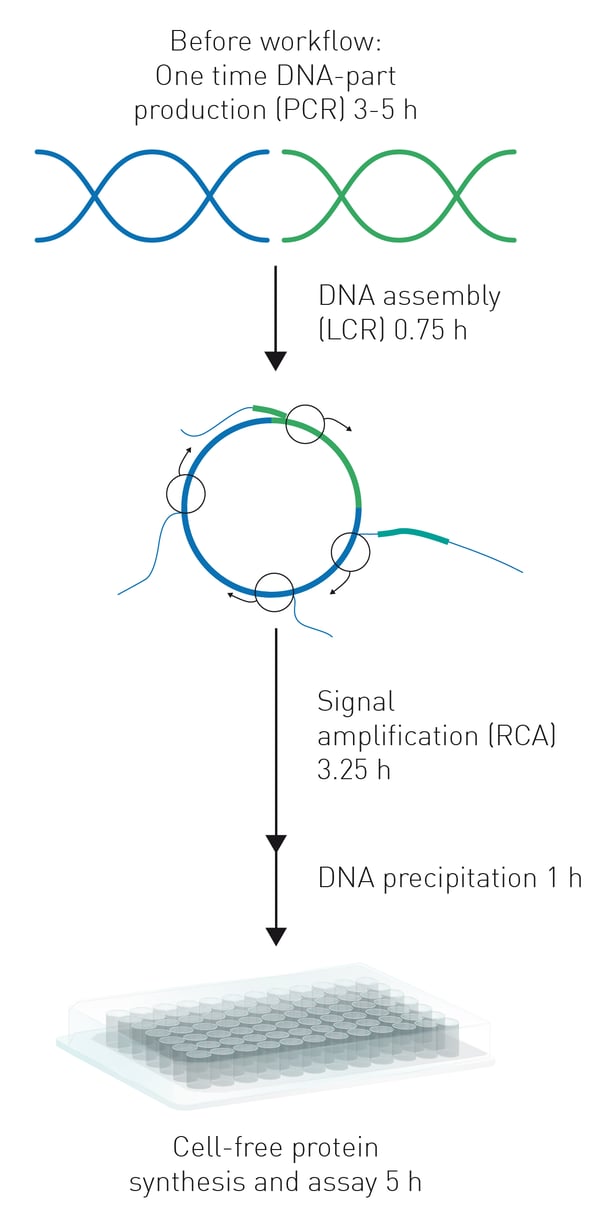
The HyperXpress workflow represents a partially automated, successive single vessel reaction within 384-well plates. It is started with a ligase cycling reaction (LCR) which mediates a non-homologous assembly of selected DNA fragments to form a circular product (figure 1). This product is then amplified in the subsequent RCA to form complex double strand DNA concatamers which serve as the templates for the gene expression and protein synthesis in cell-free extracts. In this example GFP was used as target for mutagenesis and functionality of new GFP variants was tested by comparing their fluorescence properties to the unmodified sfGFP template using a CLARIOstar® Plus microplate reader.
- Microplate (skirted 384-well PCR Plate clear, VWR)
- BMG LABTECH CLARIOstar® Plus
- Thermocycler (Tetrad2 with 384-well block, Bio-Rad)
Ligase cycling reaction
5’-phosporylated DNA fragments produced by PCR were ligated in a LCR. For this, a LCR mixture containing DNA fragments and Ampligase® (Lucigen) in LCR reaction buffer (total volume 0.612 µL) were transferred to the wells of a 384-well plate. The plate was covered with an aluminum cover film and subjected to a LCR temperature program in a thermocycler.4
Rolling circle amplification & DNA precipitation
Circular DNA constructs were then amplified by RCA. For this, first annealing mixtures were added to each LCR sample followed by RCA mixtures after the annealing program was concluded increasing the volume to a total of 1.8 µL. RCA was performed for 3 h at 40 °C and subsequent inactivation of DNA polymerase at 65 °C for 10 min. Afterwards, the amplified DNA constructs were precipitated, washed, and resolved in 1.8 µL MiliQ water at 50 °C for 10 min.
Cell-free extract protein synthesis & fluorescence assay
Subsequently, RCA-amplified DNA templates were used for the synthesis of GFP variants in cell-free extracts by adding 1.8 µL of cell-free extract solution to each well, consisting of cell-free extract from E.coli cells and cell extract buffer. The resulting 3.6 µL cell-free extract synthesis mixtures were incubated for 5 h at 29°C in the CLARIOstar Plus.
For this, the 384-well microplate was covered with an optical adhesive cover. Protein synthesis was monitored by kinetic fluorescence measurements throughout the incubation period.
Instrument settings
|
Optic settings
|
Fluorescence, plate mode, top optic
|
|
|
Monochromator settings
|
Excitation 470-7.5 |
|
| General settings |
Number of flashes | 20 |
| Settling time | 0.1 sec | |
|
Kinetic settings
|
Number of cycles
|
61
|
|
Cycle time
|
300 sec
|
|
|
Gain
|
150
|
|
Results & Discussion
To optimise their novel cell-free extract protein synthesis workflow, the authors first performed comparative measurements using a GFP expression construct. Results showed that usage of the complete workflow allowed the production of increasing amounts of functional GFP over time, while synthesis in cell-free extracts without prior amplification of DNA by RCA or with a negative control construct resulted in no GFP production (figure 2). Additionally, dialysis of RCA amplified DNA was examined. Even though dialysis led to a nearly 1.5-fold increase in total fluorescence signal, it was excluded from the final workflow since it would circumvent the one-vessel approach.
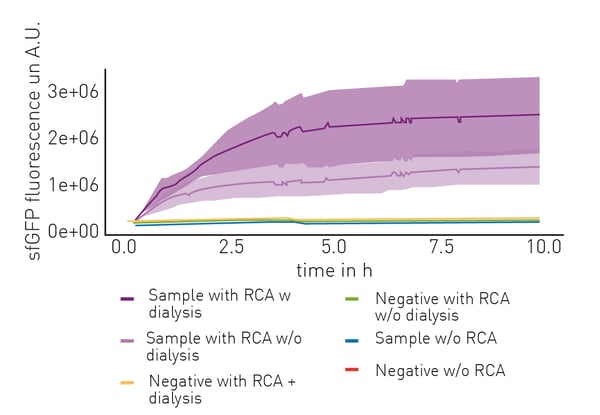 In turn, both total work volume and cost of reagents were reduced by using a self-made RCA solution. Furthermore, to ensure that reactions without GFP fluorescence did not result from a failed LCR, mKate2 co-expression was included as a control reporter.
In turn, both total work volume and cost of reagents were reduced by using a self-made RCA solution. Furthermore, to ensure that reactions without GFP fluorescence did not result from a failed LCR, mKate2 co-expression was included as a control reporter.
Based on this optimised workflow, LCR-based non-homologous recombinations between the GFP wild types sfGFP (G), mAvicFP1 (A), and mNeonGreen (N) were carried out. This included the semi-rational insertion, deletion, and substitution of β-strands of the GFP-β-barrel. For this purpose, each GFP wild-type was divided into 7 fragments based on amino acid sequence homologies, while also making sure that all fragments carry complete secondary structural elements.
Among the 12 substitution and 12 insertion variants, nine new-to-nature variants with GFP fluorescence activity were identified using the HyperXpress cell-extract workflow (figure 3). While four of these needed to be categorized as mKate2 negative, in vivo expression confirmed them as novel functional GFP variants.4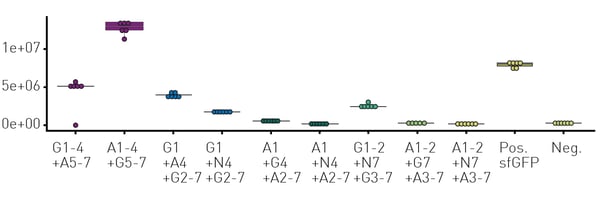
Conclusion
As demonstrated in this application note, protein prototyping and testing can be rapidly sped up by using protein synthesis in cell-free extracts. The presented HyperXpress workflow enables rapid prototyping of novel protein variants in a single vessel in vitro approach, combining flexible and combinatorial DNA assembly using LCR, the selection and amplification of circularised DNA by RCA and the protein synthesis in cell-free extracts. And all this in a down-scaled low µL approach, which allows a batch consistent and affordable throughput of thousands of samples. The CLARIOstar Plus microplate reader was the ideal measurement platform for the evaluation of the HyperXpress workflow due to its sensitivity, incubation capacity and flexibility offered by its LVF monochromator™.
References
-
Cole, M.F. and Gaucher, E.A. Exploiting Models of Molecular Evolution to Efficiently Direct Protein Engineering. Journal of Molecular Evolution (2011), 72, 193-203.
-
Nannemann, D.P., et al. Assessing directed evolution methods for the generation of biosynthetic enzymes with potential in drug biosynthesis. Future Medicinal Chemistry (2011), 3, 809-819.
-
Chen, F., et al. Reconstructed evolutionary adaptive paths give polymerases accepting reversible terminators for sequencing and SNP detection. Proceedings of the National Academy of Sciences (2010), 107, 1948-1953.
-
Zibulski, D.L., et al. HyperXpress: rapid single vessel DNA assembly and protein production in microliterscale. Front Bioeng Biotechnol (2022), 10: 832176