Introduction
Most cell-based assays rely on a single measurement yielding limited information about the biological system being studied. Multiplexed reporter assays seek to address this limitation. Particularly, luciferases have advantages for multiplexing over fluorescent proteins, including higher sensitivity, wider dynamic range and the absence of auto-luminescence. The most common multiplex luciferase assay (the dual reporter assay) uses sequential detection of two luciferases with distinct substrate specificity and the ability to quench the first luciferase. Here we describe how the expanding palette of
luciferases can now be employed such that 6 signals may be discriminated by adding spectral deconvolution.
Assay Principle
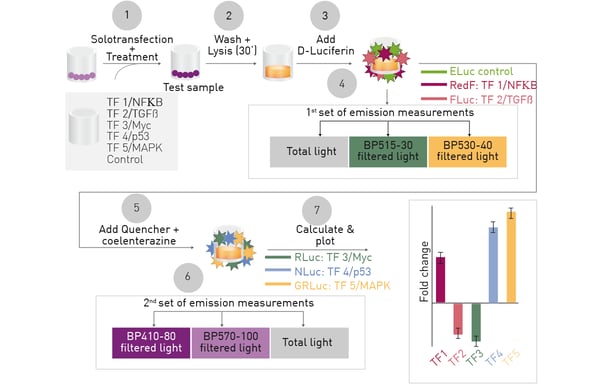
The essential aspects of the multiplex hextuple luciferase assay are shown in Figure 1.
One vector encoding six cellular signaling reporters, each associated with a distinct luciferase is solotransfected into cells. After incubation to allow luciferase expression, cells can be treated and total luciferase signals, or filtered light, detected for 3 D-Luciferin (D-LUC)-consuming luciferases followed by quenching and similar detection of 3 coelenterazine (CLZN)-consuming luciferases. Overlapping spectra are deconvoluted through calculation so that the relative expression for each signaling reporter can be found.
Materials & Methods
- Cellstar white, 384-well microplates (Greiner Bio-One)
- Dual Luciferase® Reporter (DLRTM) Assay (Promega)
- CLARIOstar®
- Luciferases (Addgene)
Experimental Procedure
Plasmid preparation, cell culture and plasmid transfection were performed as described1-5. Luciferase emissions were obtained using DLRTM reagents 24 hours after transfection with or without treatments. RNAi was performed for 24 hours before transfection. Cells were washed with PBS and lysed with Passive Lysis Buffer and 5 µl of lysate was transferred to microplates for data collection.
Measurement of Unique Bioluminescent Spectra
Luciferase measurements were initiated using LARII buffer for D-LUC-consuming luciferases or LARII and Stop & Glo® buffer for CLZN-consuming luciferases. They were read on a CLARIOstar using spectral scanning for each luciferase individually first to find cohorts that could be distinguished from one another while using the same substrate.
Instrument Settings
|
Optic settings
|
Luminescence, spectral scanning, endpoint
|
|
|
LVF settings
|
Scan wavelength 350 to 700 nm
|
|
|
Resolution 1 nm steps
|
||
|
Bandpass 10 nm
|
||
|
General settings
|
Settling time
|
0s
|
|
Measurement interval time
|
1 s
|
|
|
10 µl (followed by 5 min delay after substrate addition)
|
||
Measurement of Multiplex Bioluminescent Spectra
Luciferase measurements were obtained on a CLARIOstar using LARII buffer for D-Luc turnover then Stop & Glo® buffer injections and the following settings for D-LUC- and CLZN-consuming luciferases.
|
Optic settings
|
Luminescence, well mode kinetic, multichromatic
|
|
||||
|
Filters
|
D-LUC
|
Lens
|
515-30
|
530-40
|
|
|
|
CLZN
|
410-80
|
570-100
|
Lens
|
|
||
|
General settings
|
Measurement interval time
|
2 (D-LUC) or 1 (CLZN) sec
|
|
|||
|
Settling time
|
0.1 s
|
|
||||
|
Kinetic settings
|
Number of cycles
|
2
|
|
|||
|
Cycle time
|
11.54 (D-LUC) or 11.72 (CLZN) s
|
|
||||
|
Reagent injection
|
10 µl (followed by 30 s delay: D-Luc)
15 µl (followed by 7 s delay: CLZN) |
|
||||
Results & Discussion
Twelve luciferases were characterized to determine the ability to spectrally resolve multiple signals. Figure 2 shows the results for 6 D-LUC- (Left) and 6 CLZN-consuming luciferases (Right)1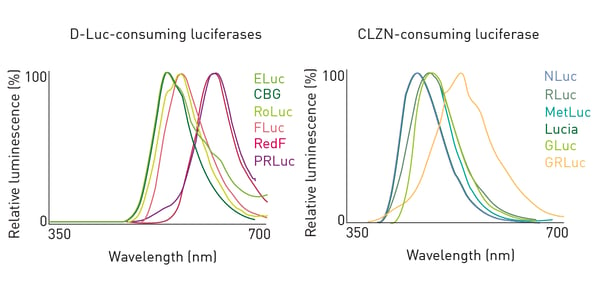 Based on the results of the above spectral data as well as substrate specificity and quenching capacity1, 6 luciferases were brought forward for further testing (Table 1).
Based on the results of the above spectral data as well as substrate specificity and quenching capacity1, 6 luciferases were brought forward for further testing (Table 1).
Table 1: Luciferases used in hextuple multiplexing (* results indicate that CLZN is a suitable substrate for NLuc)1
| Abbreviation | Full name | Emission peak | Substrate |
| ELuc | Enhanced Beetle Luciferase | 537 nm | D-Luc |
| FLuc | Firefly Luciferase | 562 nm | |
| RedF | Red Firefly Luciferase | 614 nm | |
| NLuc* | Nano Luciferase | 462 nm | CLZN |
| RLuc | Renilla Luciferase | 481 nm | |
| GRLuc | Green Renilla Luciferase | 532 nm |
These 2 groups of 3 luciferases each were each further characterized to ensure that the contribution of each luciferase could be deconvoluted from the light produced by all, based on the filters shown in instrument settings and mathematical calculations1. In addition, assessment of signal stability showed that the delay after injection, multichromatic order and integration measurement time in the instrument settings are suitable1. Furthermore, experiments showed that the experimental approach employed allows for accurate quantification of static and dynamic levels of each luciferase in a mixture1.
Proof of principle testing was performed to detect direct and collateral effects of knocking down a single constituent of the multiplex luciferase assay by proven siRNA approaches (Figure 3). Besides target knockdown, clear effects on the expression of other constituents may be observed.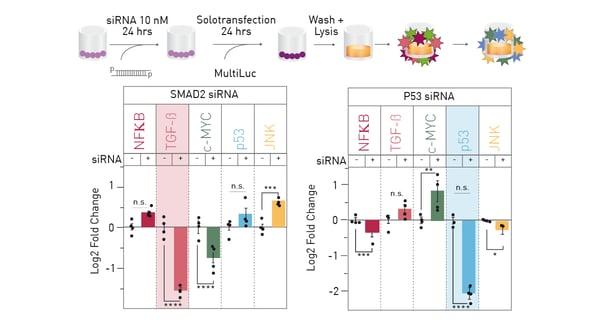
Conclusion
The CLARIOstar microplate reader proved to be the ideal tool for developing this assay. Using spectral scanning, multiple luciferases could be characterized to find cohorts that could be distinguished from one another. Subsequently, the combined use of multichromatics, emission filters and reagent injectors enabled the establishment of a complete multiplex hextuple luciferase assay detection system.
Multiplex hextuple luciferase assaying greatly expands our ability to assess the effects of siRNA, ligand, and chemical compound treatments. This system provides the data-rich approach necessary for a holistic understanding of complex biological systems.
References
- Sarrion-Perdigones et al. Examining multiple cellular pathways at once using multiplex luciferase assaying. Nat. Commun. (2019) 10: 5710.
- Sarrion-Perdigones et al. Rapid and efficient synthetic assembly of multiplex luciferase reporter plasmids for the simultaneous monitoring of up to six cellular signaling pathways. Curr. Protoc. Mol. Biol. (2021) 131: e121.
- Sarrion-Perdigones et al. Simultaneous examination of cellular pathways using multiplex hextuple luciferase assaying. Curr. Protoc. Mol. Biol. (2021) 131: e122.
- Synthetic assembly DNA cloning of multiplex hextuple luciferase reporter plasmids. Methods Mol. Biol. (2022) 2524: 409-432.
- Sarrion-Perdigones et al. Multiplex hextuple luciferase assaying. Methods Mol. Biol. (2022) 2524: 433-456.