Introduction
One of the critical steps in drug development is the assessment of binary complex formation between compounds and their target proteins of interest (POI). Usually, binary complex formation is investigated by testing purified proteins in vitro using fluorescence polarization, isothermal titration calorimetry (ITC), surface plasmon resonance (SPR) or differential scanning fluorimetry (DSF) methods.1 However, most proteins need to be truncated for soluble expression and assays like ITC require high protein concentrations or immobilisation of the protein as found in SPR. Fluorescence polarization on the other hand is a tracer-based method requiring a binder excluding the discovery of novel binders. While DSF does not require a tracer it highly interferes with coloured compounds (especially red-yellow) severely restricting the amount of compatible test samples. Accordingly, there is a need for experimental setups that can improve the screening for new ligands.
One possible approach is the Cellular Thermal Shift Assay (CETSA), which allows the measurement of endogenous full-length protein in a tracer-free manner, however normally requires subsequent proteomics or antibody-based detection which drastically decreases the throughput.2 Herein, the authors describe a HiBiT CETSA which addresses this problem. For this, the POI´s gene locus is modified to express a HiBiT fusion tag, circumventing the needs for specific antibodies or to measure proteomics which allows to run HiBiT CETSA in a target-focused system.3
The HiBiT CETSA enables the qualities of DSF, which allows novel binder identification with low amounts of protein, without the downsides in form of protein truncations and restrictions in compound selection.4 HiBiT CETSA therefore allows screening efforts including proteins which are not available in vitro.
Assay principle
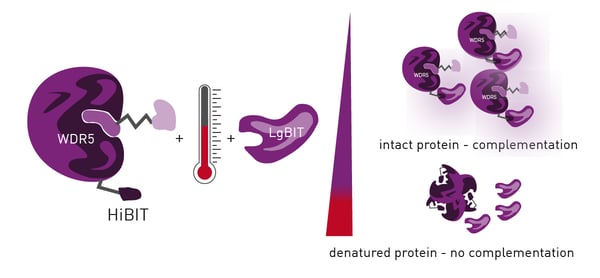
HiBiT CETSA identifies interactions between a POI and potential ligands by determining a POI`s melting point along a heat gradient. Addition of ligands to the POI can increase its stability, leading to a thermal shift of its melting point.
HiBiT CETSA was developed to increase the throughput of CETSA to 384- or 1536-well formats with an endpoint readout. For this purpose, HiBiT-tagged POI is endogenously expressed by the cells which are treated with compounds. After incubation along a temperature gradient, the cells are lysed for LgBiT complementation which results in a luminescence signal. Temperatures exceeding the POI melting point cause POI aggregates which disable LgBiT complementation (figure 1). Plotting the temperature against the detected luminescence allows to visualise melting curves and identify compound-induced thermal shifts by calculating their IC50 values.
Materials & Methods
- 96-well PCR plates (STARLAB; I1402-9800)
- Gradient PCR cycler (Biometra)
- White 384-well LDV plates (Greiner; 784075)
- Cell line with endogenous HiBiT-tagging (Here, HEK293T HiBiT-WDR5)
- PHERAstar® FSX (BMG LABTECH)
- HiBiT lytic detection reagent (Promega)
- PROTACs were previously synthesised4
- OptiMEM medium (Gibco)
Experimental procedure
Cell preparation and temperature exposure
HEK293THiBiT-WDR5 were used in a concentration of 200,000 cells/ml and resuspended in OptiMEM medium. The cells were incubated with 10 μM of the corresponding compound for 1 h at 37 °C and 5% CO2. Incubated cells were transferred into 96-well PCR plates and incubated for 3 min in a gradient PCR cycler, followed by 3 min cool down at 25 °C. 10 μl of the incubated cells were transferred into white 384-well LDV plates and 10 μl HiBiT lytic detection reagent was added. Readout was carried out after 10 min incubation at room temperature in a PHERAstar FSX plate reader using the LUM plus optical module.
Instrument settings
|
Optic settings
|
Luminescence, plate mode kinetic
|
|
|
Filter
|
LUM plus |
|
| Incubation |
23 °C
|
|
|
General settings
|
Settling time
|
0 s
|
|
Measurement interval time
|
0.26 s
|
|
Results & Discussion
To establish the HiBiT CETSA, 3 min incubation and direct cool down in the thermocycler to 25 °C was shown to be the best protocol for sample treatment. Subsequently, the lysis and detection steps were tested for the optimal readout time since lysis, complementation and NanoLuc intensity are time dependent. To analyse the optimal signal to noise ratio and to prove the independency of the melting IC50 to the signal intensity, cells treated with DMSO and the control compound OICR-9429 were measured for 10 min.
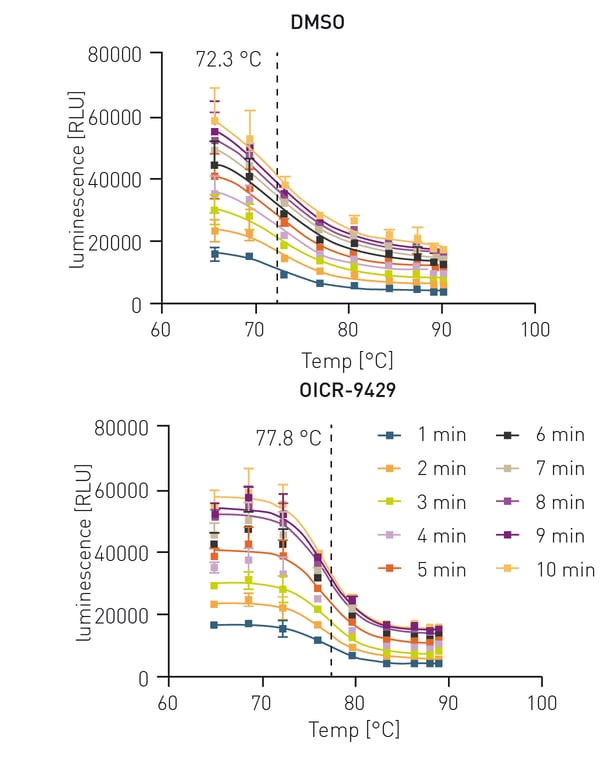 Since the assay signal increased drastically with time (figure 2) but still maintained its IC50, a 10 min incubation was chosen. Next, a set of diverse WDR5 targeting PROTACs were measured in this assay to generate binding data towards WDR5.
Since the assay signal increased drastically with time (figure 2) but still maintained its IC50, a 10 min incubation was chosen. Next, a set of diverse WDR5 targeting PROTACs were measured in this assay to generate binding data towards WDR5.
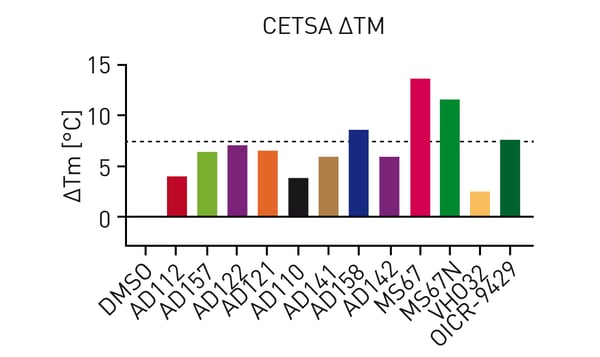
Addition of OICR-9429 to the HiBiT CETSA resulted in significant thermal shifts of up to 7.5 °C (figure 3). Additionally, the negative control VH032 (VHL ligand) resulted in a similar melting point as DMSO indicating assay robustness. Subsequently, thermal shifts for a set of OICR-9429-based compounds were collected (figure 4). Comparison of the measured compounds showed a crowding of all PROTACs from the AD series around the reference (OICR-9429) while the optimised WDR5 binders MS67 and MS67N showed a significantly higher melting point as expected. Additionally, the influence of compound linker length on binding could be observed. While AD112 (no linker) shows a negative influence of E3 handle attachments, increasing length of aliphatic linkers increases the binding towards the parent compound (AD157=C3, AD122=C4). However, even longer linkers (AD110=C6) show a decrease in affinity towards WDR5.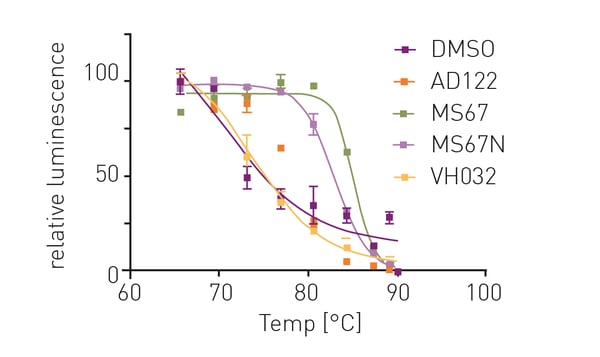
Conclusion
The HiBiT CETSA was successfully established in 10 µl reactions using the PHERAstar FSX with its high sensitivity. The HiBiT CETSA´s small assay volumes reduce reagent use and dramatically increase assay throughput providing significant cost savings. Further downscaling to 1536-well plates is possible for proteins with sufficient expression levels and was also successfully detected in the PHERAstar FSX plate reader. The tracer-free HiBiT CETSA was shown to be the superior target engagement assay for cellular WDR5 compared to tracer-based methods highlighting the potential for drug discovery of HiBiT CETSA.4
For more information on these kinds of experiments and if you are interested in more ways to evaluate PROTACs and other protein degrader check out this scientific talk.
References
-
Schwalm, MP, et al. A Toolbox for the Generation of Chemical Probes for Baculovirus IAP Repeat Containing Proteins. Frontiers in Cell and Developmental Biology 10 (2022).
-
Jensen, AJ., et al. CETSA: a target engagement assay with potential to transform drug discovery. Future medicinal chemistry 7.8 (2015): 975-978.
-
Mortison, JD., et al. Rapid evaluation of small molecule cellular target engagement with a luminescent thermal shift assay. ACS Medicinal Chemistry Letters 12.8 (2021): 1288-1294.
-
Schwalm, MP., et al. Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization. Cell Chem Biol., 2023, S2451-9456(23)