Introduction
In recent years, YEATS domains have joined the ranks of epigenetic regulators and are recognised as bona fide readers of histone post-translational modifications (HPTM) alongside bromodomains, PHD fingers, and others. YEATS domains bind to lysine when the ε-carbon is acetylated or crotonylated1. In contrast to acetylation, the significance of crotonylation in active translation is poorly understood2. The YEATS-containing ENL has been confirmed as a major driver of several types of acute leukaemia and links histone acetylation to active transcription (Fig. 1). Further to this, CRISPR/Cas9-mediated knockdown of ENL reduces malignant behaviour in leukaemic cell lines, while its intact YEATS domain promotes leukaemic growth3,4. These observations highlight ENL as a rational drug target to attenuate aberrant cell growth and malignancy and it is therefore a target of ongoing drug discovery programmes.
Here, we present the development of a selective inhibitor of the YEATS domain of ENL. We aspire to develop chemical probes for each member of the YEATS family that can be used as tool compounds to interrogate their biological function.
Assay Principle
We used the PerkinElmer AlphaScreen® Histidine Detection Kit that consists of streptavidin coated “donor” beads and Nickel chelator coated “acceptor” beads. Excitation of the donor beads with light at 680 nm drives the conversion of oxygen in solution to excited singlet oxygen (1O2). Acceptor beads convert the energy of the singlet oxygen to light between 520 and 620 nm (Fig. 2). Due to the short half-life of singlet oxygen, donor and acceptor beads must be in proximity for successful energy transfer. Coupling the donor bead via Streptavidin to acylated histone 3 and the acceptor bead to the YEATS domain, both beads are brought together when protein and peptide interact. Disrupting the protein-peptide interaction (and therefore increasing the distance between the associated donor and acceptor beads), reduces the signal and provides a measurable output.
Materials & Methods
- PHERAstar FS and FSX with Stacker (BMG LABTECH)
- Labcyte® Echo® liquid handler
- AlphaScreen® Histidine Detection Kit (PerkinElmer) Acylated and biotinylated histone 3 derived peptides (LifeTein)
- 6His tagged YEATS domains of ENL, AF9, YEATS2 and GAS41 (produced in house)
- 384 well ProxiPlates (Perkin Elmer), assay volume was 20 µl
Instrument settings
| Optic settings | AlphaScreen, endpoint |
|
| Optic Module: | AlphaScreen 680 570 | |
| Gain: | 3500 | |
| General settings |
Settling time: | 0.0 s |
| Excitation time: | 0.06 s | |
| Integration start: | 0.09 s |
|
| Integration time: | 0.20 s | |
Experimental procedure
Compounds were dispensed into the plates using the Echo® liquid handler (final assay concentration of 50 µM for single shot experiments, top concentration of 200 µM for dose response curves).
Protein, peptide and compound were incubated for 30 min before the addition of AlphaScreen® beads (1:600). The plates were then incubated for at least 60 min before being read.
A counter-screen with a [biotin]-His6 peptide instead of a [biotin]-peptide:YEATS-His6 pair was also performed to identify and therefore exclude false positives (e.g. metal chelators or fluorescence quenchers).
Results & Discussion
Assay optimisation
To determine the best assay conditions, we first performed a protein versus peptide titration for each protein prep. The molar ratio of protein to peptide from which a small deviation would not lead to a marked change in luminescence signal was then chosen for the screening campaign (Fig. 3).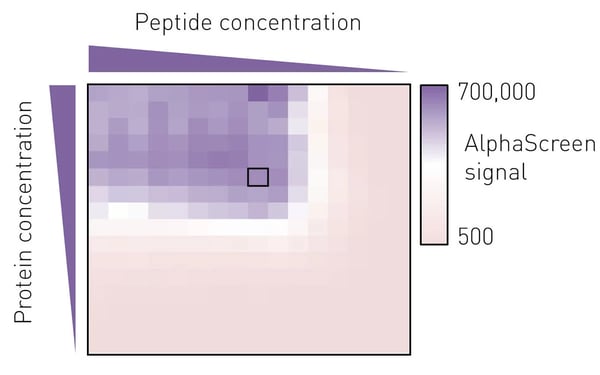
Screening campaign
Several libraries (including an in-house bromodomain inhibitor library, and the 16k and 24k compound libraries of the Ontario Institute for Cancer Research) were screened against ENL in the first instance as ‘single shots‘ (50 µM final assay concentration, in duplicate). Compounds that showed inhibition here were retested at lower concentrations to establish potency. Of these, compounds showing >80% inhibition below 50 µM were carried forward for full concentration-response curves with all four human YEATS domains (ENL, AF9, YEATS2, GAS41), again using the AlphaScreen® assay. For the most potent compound, IC50 values were obtained for ENL:H3K18ac (2.7 µM) and AF9:H3K9ac (7.3 µM) while no IC50 values could be determined for YEATS2A:H3K27cro and GAS41:H3K9ac in the concentration range tested (Fig. 4).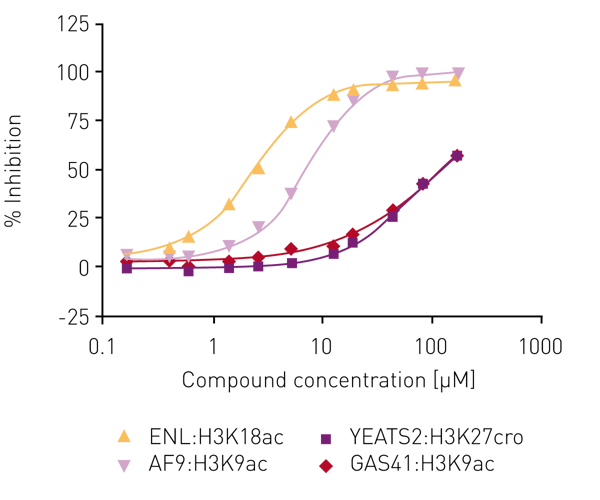
Conclusion
We have identified a potent small molecule inhibitor of the closely related ENL and AF9 that demonstrates selectivity over the other two human YEATS domain-containing proteins (YEATS2 and GAS41). The results of the original AlphaScreen® screening campaign are in good agreement with the orthogonal assays (data not shown).
The PHERAstar FSX provides a powerful and versatile platform for drug discovery campaigns. Here, the screening of tens of thousands of compounds for inhibiting the YEATS domain with an AlphaScreen® approach resulted the identification of a potent small molecule inhibitor.
References
- Y. Li, H. Wen et al. (2014) AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell, vol. 159, no. 3, pp. 558–571
- A. Dutta, S. M. Abmayr, and J. L. Workman (2016) Diverse Activities of Histone Acylations Connect Metabolism to Chromatin Function. Mol. Cell, vol. 63, no. 4, pp. 547–552
- M. A. Erb et al. (2017) Transcription control by the ENL YEATS domain in acute leukaemia. Nature
- L. Wan et al. (2017) ENL links histoneacetylation to oncogenic gene expression in acute myeloid leukaemia. Nature


