
PHERAstar FSX
Powerful and most sensitive HTS plate reader
Life in the depths of the ocean operates under extreme conditions. Find out how proteins from deep-sea luminescent organisms are useful for measurements on microplate readers.
 Dr Barry Whyte
Dr Barry Whyte
Our knowledge of the deep-sea, the largest biome on Earth, is in its infancy. Water covers more than 70% of the planet and the deep-sea accounts for more than 300 million km2 of the Earth’s surface. 1,2 Scientists have only scratched the surface of life in the ocean depths but there are intriguing benefits to studying its organisms and ecosystems. In this blog, we look at some of the bioluminescent tools that have been developed from proteins isolated from luminescent organisms flourishing in these environments.

 The usefulness of bioluminescence as a tool to study biology at the molecular level jump started a search for alternative luciferases, in many cases from other marine organisms that were bioluminescent. But what makes the enzymes from marine organisms particularly attractive as molecular probes?
The usefulness of bioluminescence as a tool to study biology at the molecular level jump started a search for alternative luciferases, in many cases from other marine organisms that were bioluminescent. But what makes the enzymes from marine organisms particularly attractive as molecular probes?Bioluminescence occurs when luciferase enzymes oxidize the photon-emitting substrate luciferin. Luciferins typically react with molecular oxygen. The net result of the enzyme-catalyzed reaction is an intermediate of the substrate luciferin in an excited state that emits light upon decaying to its ground state.
If you were to select the ideal luciferase as a starting point for engineering a new molecular probe, it might include one that has high sensitivity, bright signal, and small size. These qualities make luciferase suitable for a wide range of applications, nimble enough for example to study protein-protein interactions, protein-small molecule interactions, gene responses or for performing compound screening.
The GLuc and OLuc luciferases from G. princeps and O. gracilirostris are significantly smaller in size (20- and 19-kDa, respectively) compared with Renilla and firefly luciferase. This property alone has prompted researchers to try and engineer them for optimal reporter qualities.
Inspired by the small size of OLuc-19, a team of scientists set out on the task to engineer a better luciferase. 3,6 The ideal enzyme should be small, monomeric and structurally stable to environmental conditions. Recombinant OLuc-19 while small in size did not retain many of the desirable qualities of its larger native enzyme.
The researchers initiated a three-step program to put this straight: a random mutagenesis step to screen for brighter luminescence; identification of a better substrate to further increase brightness; and screening using the new substrate of another random library of mutants to find even brighter bioluminescence. The endpoint for this work was the engineered luciferase NanoLuc, an enzyme with a novel substrate furimazine and improved physical and chemical features up to 100 times brighter than its Renilla and firefly luciferase counterparts.
What types of applications are possible with NanoLuc and other luciferases and how can microplate readers be used for this type of research?
In the application note NanoBRET assay for monitoring of ligand binding to GPCRs in live cells, using the CLARIOstar and the PHERAstar FS, NanoLuc was used together with an acceptor fluorophore in a NanoBRET™ or Nano Bioluminescence Resonance Energy Transfer assay to monitor for ligand binding to G-protein coupled receptors (GPCRs). GPCRs play a crucial role in the human body and many other organisms. They serve as gateways into the cell for a range of external signals, which include hormones, neurotransmitters, and other environmental stimuli. These signals lead to a cascade of intracellular signalling events that trigger downstream processes essential for ensuring the vitality of the cell. You can read more in the blog G protein-coupled receptors.
In the specific assay described in this application note, a GPCR was expressed in live HEK293 cells with NanoLuc at the N-terminus. The receptor ligand was labelled with a different fluorophore suitable for BRET in proximity-based interactions. Only blue emission of NanoLuc is detected if the ligand is not bound to the receptor. In contrast, red fluorescence is additionally detected if the ligand binds to the receptor due to excitation of the fluorescence label by resonance energy transfer from NanoLuc. The tests were performed on the CLARIOstar® Plus or PHERAstar® FSX.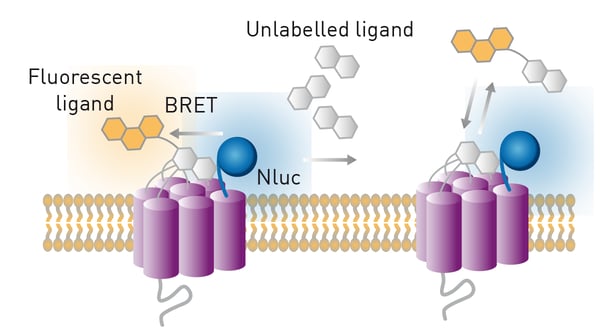 The luciferase NanoBRET system was used in competitive binding experiments involving modified receptors and different fluorescently labelled ligands to determine IC50 and KD values. NanoBRET was shown to be a viable alternative to existing methods for the determination of ligand binding to GPCRs. It has the advantage of being a high-throughput method that does not require the use of radioactivity.
The luciferase NanoBRET system was used in competitive binding experiments involving modified receptors and different fluorescently labelled ligands to determine IC50 and KD values. NanoBRET was shown to be a viable alternative to existing methods for the determination of ligand binding to GPCRs. It has the advantage of being a high-throughput method that does not require the use of radioactivity.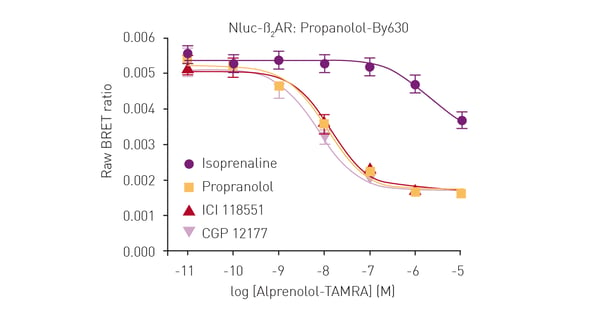 Luciferases also find applications in determining the genotoxic properties of different chemicals. This type of screening is essential to ensure the safety of pharmaceuticals, industrial chemicals and products like those used for personal care.
Luciferases also find applications in determining the genotoxic properties of different chemicals. This type of screening is essential to ensure the safety of pharmaceuticals, industrial chemicals and products like those used for personal care.
The application note BlueScreen HC - a luminescence based, high-throughput, in vitro genotoxicity assay describes the use of the GLuc luciferase from G. princeps to measure genotoxicity. In this luciferase assay, exposure to a genotoxic chemical causes increased expression of the GADD45a gene which is coupled to the luciferase reporter system. GADD45a stands for growth arrest and DNA damage inducible alpha. In organisms harboring this gene, its transcript levels increase following stressful growth arrest conditions or treatment with DNA-damaging agents. These properties have been used to construct an in vitro test for mutagenicity which is at the heart of BlueScreen HC™.
BlueScreen HC™ from Gentronix is a high-throughput genotoxicity test that uses a genetically engineered TK6 cell line and is suitable for miniaturization. The cells are engineered to have the GADD45a gene coupled to the G. princeps luciferase. The amount of luciferase produced by the BlueScreen HC cell lines is assessed by injection of the substrate coelenterazine, which results in a short-lived flash luminescence signal.
![Fig.5: BlueScreen HC S9 assay positive cytotoxicity (a) and genotoxicity (b) results for 20 µg/ml benzo[a]pyrene. Error bars show ±1 standard deviation based on 4 replicate analyses on separate microplates.](https://www.bmglabtech.com/hs-fs/hubfs/1_Webseite/5_Resources/Blogs/deep-sea-blog-Fig.5.webp?width=600&height=646&name=deep-sea-blog-Fig.5.webp) Exposure to a genotoxic chemical gives a dose-dependent increase in the production of luciferase from the GADD45a reporter gene.
Exposure to a genotoxic chemical gives a dose-dependent increase in the production of luciferase from the GADD45a reporter gene.
In addition, reduced cell proliferation, a measure of cytotoxicity, was assessed either by changes in absorbance in BlueScreen HC or in fluorescence from a DNA stain in BlueScreen HC S9. Genotoxicity and cytotoxicity were measured simultaneously using flash luminescence, absorbance, and fluorescence measurements. BlueScreen HC and BlueScreen HC S9 are fully compatible with BMG Labtech microplate readers using the three detection modes and reagent injector systems. The combination of microplate reader and assay kits provided fast, accurate and reproducible results for the detection and quantification of genotoxic liability for chemicals with different potencies and modes of action.
A crucial part of making accurate and reliable luciferase measurements is the need to maintain cell health. The Atmospheric Control Unit (ACU) on the CLARIOstar is ideal for this purpose and helps maintain cells under precisely defined conditions so that long-term cell-based assays can be carried out under different experimental conditions.
In the application note The new Atmospheric Control Unit (ACU) for the CLARIOstar provides versatility in long-term cell-based assays. The CLARIOstar microplate reader and ACU was able to fully sustain the normal proliferation and health of untreated K562 cells for the entire duration of experiments. This was essential to permit the detection of time-dependent and dose-dependent effects on cell proliferation and cytotoxicity due to the tested compounds.
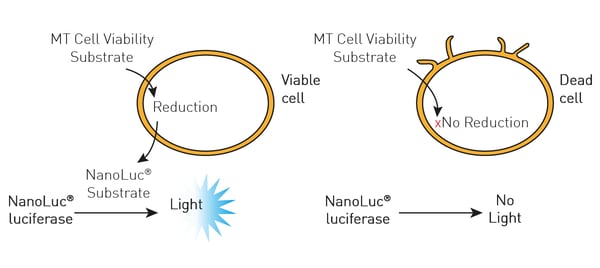 In this case, NanoLuc luciferase was used to measure cell viability (RealTime-Glo® MT Cell Viability Assay). The RealTime-Glo® MT Cell Viability Assay is a bioluminescent test that relies on the metabolic (MT) reducing potential of cells. NanoLuc luciferase and cell-permeant pro-NanoLuc® substrate were added to cells in culture. Viable cells reduce the pro-NanoLuc® substrate which diffuses into the cell medium. In the medium, the substrate is rapidly converted by NanoLuc luciferase to emit luminescence proportional to the number of viable cells.
In this case, NanoLuc luciferase was used to measure cell viability (RealTime-Glo® MT Cell Viability Assay). The RealTime-Glo® MT Cell Viability Assay is a bioluminescent test that relies on the metabolic (MT) reducing potential of cells. NanoLuc luciferase and cell-permeant pro-NanoLuc® substrate were added to cells in culture. Viable cells reduce the pro-NanoLuc® substrate which diffuses into the cell medium. In the medium, the substrate is rapidly converted by NanoLuc luciferase to emit luminescence proportional to the number of viable cells. 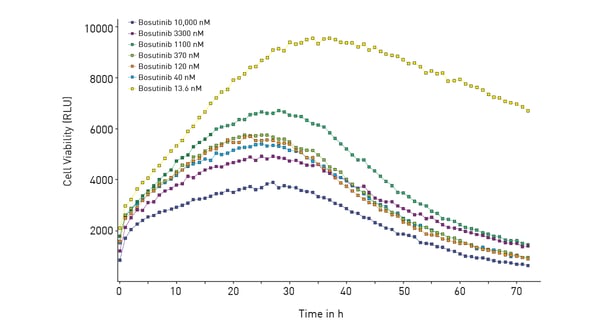 In the study, the effect of the tyrosine kinase inhibitor bosutinib was measured over time for its impact on cytotoxicity and cell viability. The ACU was shown to sustain cells for 72 hours which was the duration of the subsequent experiments with the inhibitor.
In the study, the effect of the tyrosine kinase inhibitor bosutinib was measured over time for its impact on cytotoxicity and cell viability. The ACU was shown to sustain cells for 72 hours which was the duration of the subsequent experiments with the inhibitor.
GLuc and NanoLuc like other luciferases can be used in dual luciferase reporter assays which are widely used to investigate gene transcription and regulation. Today researchers have several options to take advantage of dual-reporter formats and you can read more about the different options in the BMG LABTECH blog Wavelength based dual glow reporter genes.
BMG LABTECH microplate readers allow simultaneous dual detection and can therefore measure emission at two wavelengths at the same time. This reduces the read time in half which is a significant advantage over microplate readers that require sequential measurements. Simultaneous dual detection also helps to reduce variability due to bubbles, temperature effects and fluid movement. This makes BMG Labtech microplate readers the ideal choice for dual luciferase reporter assays.
What is the preferred BMG LABTECH microplate reader for specific needs and applications related to luminescence?
Luminescence detection can be performed on BMG LABTECH´s dedicated luminescence plate reader LUMIstar® Omega, and multi-mode microplate readers including the PHERAstar FSX, CLARIOstar Plus, VANTAstar® and FLUOstar® Omega.
All of our luminescence plate readers that are certified for Dual-Luciferase Reporter assays utilize luminescence-optimized low-noise PMT (photomultiplier tube) and can be equipped with high-precision reagent injectors.
All BMG LABTECH microplate readers have exceptionally fast reading capabilities. Collectively, these multi-mode readers combine high performance with miniaturized assays, short measurement times, and offer considerable savings on materials and other resources.
Powerful and most sensitive HTS plate reader
Most flexible Plate Reader for Assay Development
Upgradeable single and multi-mode microplate reader series
Flexible microplate reader with simplified workflows
Gene reporter assays are sensitive and specific tools to study the regulation of gene expression. Learn about the different options available, their uses, and the benefits of running these types of assays on microplate readers.
Next generation sequencing (NGS) technologies for DNA or RNA have made tremendous progress in recent years. Find out how microplate readers can advance the quality control of nucleic acids to facilitate NGS.
Measuring light scattering is an accurate and expedient way to determine drug solubility. Find out how the NEPHELOstar® Plus can be used for early drug solubility screening at high throughput.
Mitochondrial toxicity can have devastating effects on the cell and life. Find out how microplate readers can be used to assess mitochondrial health and how this impacts disease and drug discovery.
Find out how microplate readers can be used to measure histone deacetylase (HDAC) activity and assist drug discovery.