Introduction
Bioluminescence resonance energy transfer (BRET) is an established method to study protein-protein interactions. Most of the research focuses on GPCRs as these receptors play an important role in finding new drug candidates.1
There are several BRET-based approaches that allow the monitoring of protein-protein interaction2 such as:
- BRET between receptors and ligands3
- BRET between receptors and G proteins
- BRET between receptors and downstream effectors, such as adenylyl cyclase or ion channels
- BRET sensors that follow beta-arrestin recrutiment cAMP BRET biosensors
In this application we will show how a cAMP BRET biosensor can be used to monitor ligand binding with the help of the CLARIOstar microplate reader.
Assay Principle

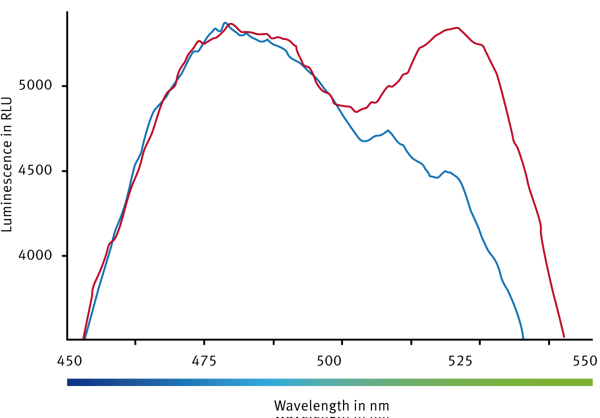
The BRET biosensor carries a cAMP binding domain, Renilla luciferase and YFP. The latter two are in close proximity. Once the luciferease is activated by substrate addition (Coelenterazine H) the enzyme emission excites YFP. This leads to a strong emission signal at 530 nm and therefore to a strong BRET signal (Fig. 1). Once a ligand is binding to a GPCR, G proteins are activated and cAMP is generated. Free cAMP will find its binding site and the biosensor will undergo a conformational change leading to a greater distance between Rluc and YFP. The result of that process is no or only weak YFP emission leading to a small BRET signal.
Once ligands are found that are able to effectuate the conformational change, a dose response can be run in order to determine EC50 values.
Materials & Methods
- White 96-well microplates from NUNC
- Coelenterazine h from Dalton Pharma Services
- CLARIOstar microplate reader from BMG LABTECH
CLARIOstar settings for ligand binding
| Detection Method: |
Luminescence, plate mode, top reading |
| Optic setting: | Well mode chromatics selected |
| Monochromator: | Just select 475-30 and 535-30 nm, the bandwidth is adjustable up to 100 nm |
| Filters: | Emission filters 475-30 and 535-30 nm |
| Kinetic settings: | No. of cycles: 10 |
| Cycle time: 90 sec | |
| Measurement time: 0.50 sec | |
| Gain: | 3600 for both channels |
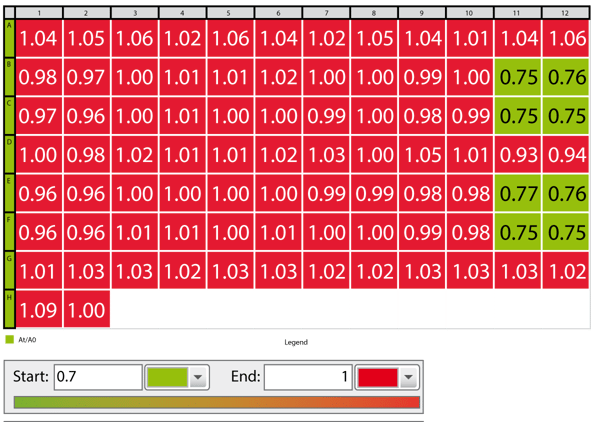
BRET calculation
- Ratio calculation: 535 nm / 475 nm
- Average of 4 first measurement points (based on ratio) = A0
- Average of 5 last measurement points (based on ratio) = At
- Divide At/A0
- Use color gradient to see positive results immediately (an example is shown in Fig. 3)
Results & Discussion
Initial BRET luminescence scan
Two controls were prepared. One well contained the biosensor without ligand – so a high BRET signal is expected (BRETMax Control). A second well consisted of the cAMP biosensor accompanied by a high ligand concentration. It was a ligand chosen that is known to lead to an increase in cAMP levels and therefore into a conformational change. For the latter a minimal BRET signal is expected (BRETMin Control). Overlayed luminescence spectra are shown in Fig. 2.
Different ligands can be tested on one microplate. A color gradient was applied in the MARS Data Analysis software to get a fast overview of wells in which the ligand was effectively binding (Fig. 3).
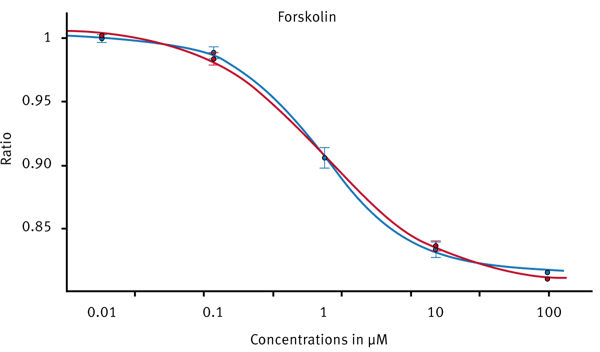
Only forskolin treated samples showed a decrease in BRET ratio. Forskolin is known to directly activate adenylate cyclase which in turn produces cAMP. Therefore, forskolin can be used as a control substance for this assay. A dose response curve was created by adding varying amounts of forskolin in replicate wells (Fig. 4)
Although the intrinsic assay window is quite small, the CLARIOstar is sensitive enough to detect small luminescence changes by either using BRET specific emission filters or by using the LVF monochromator. The standard deviation in replicates is small enough to obtain excellent 4-parameter dose response curves with R2> 0.999.
Conclusion
The CLARIOstar microplate reader is a suitable instrument to detect protein ligand binding with the help of the cAMP BRET biosensor technology. The MARS Data Analysis software can be set up to use a color gradient to fast identify ligands that lead to a receptor conformational change.
References
- Kocan and Pfleger (2011) Study of GPCR-protein interactions by BRET. Methods Mol. Biol. 746: 357-371.
- Salahpour et al. (2012) BRET biosensors to study GPCR biology, pharmacology, and signal transduction. Front Endocrinol. (Lausanne) 3:article105.
- Stoddart et al. (2015) Application of BRET to monitor ligand binding to GPCRs. Nat. Methods 12(7): 661-663.